A collection of electronic tools to help you find your way in the RNA world
Our mission
Discover new biological classes of trans RNA-RNA interactions.
Investigate their functional roles, in particular during infection in a host-pathogen settings.
Identify targets for therapeutic applications that prevent or enable particular trans RNA-RNA interactions.
Our computational methods
- Input: information on cis and/or trans interacting sequence positions and - optionally - corresponding multiple sequence alignment(s).
- Output: arc-plot
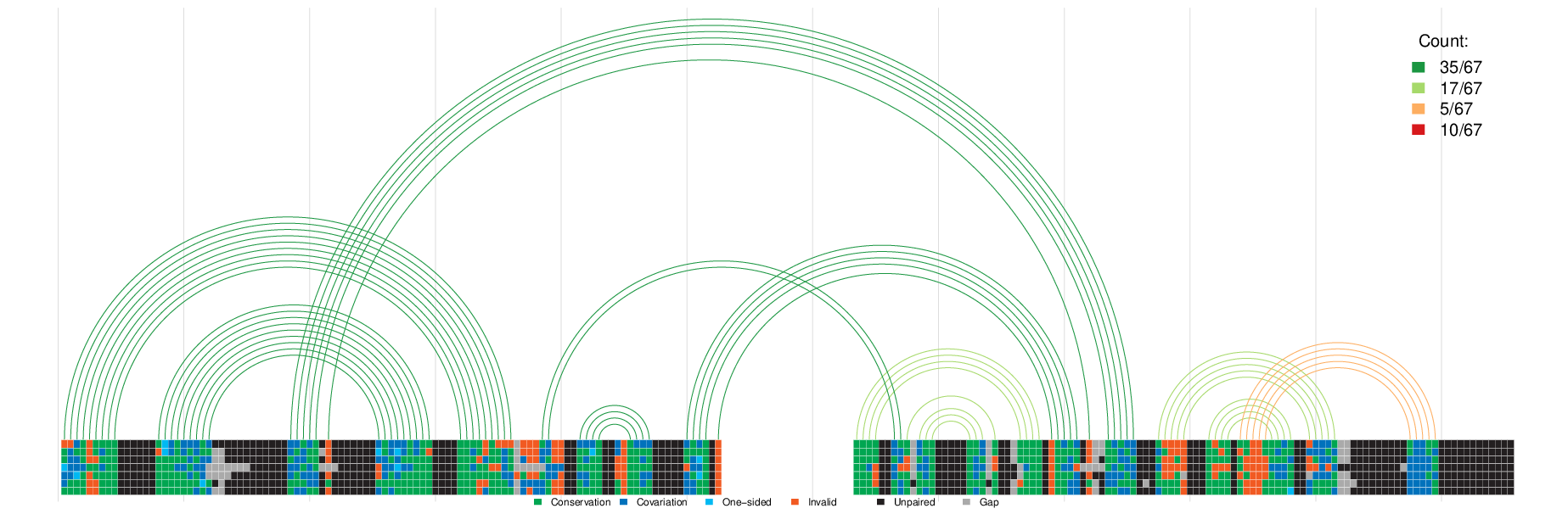
- Input: output files of CoBold
- Output: arc-plot visualising known and predicted transient RNA structure features
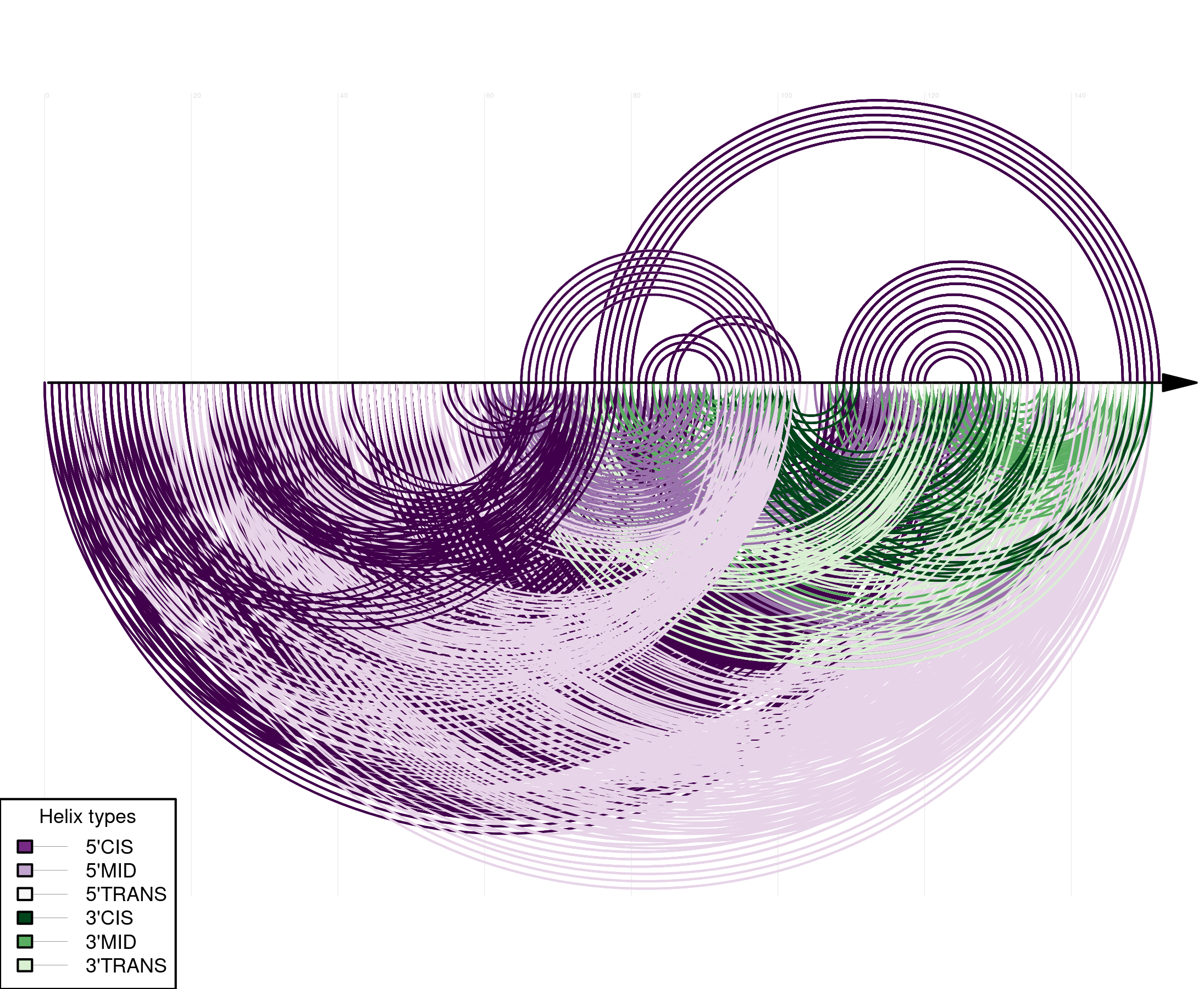
- Input: multiple sequence alignment and - optionally - a corresponding phylogenetic tree
- Output: predicted RNA secondary structure features
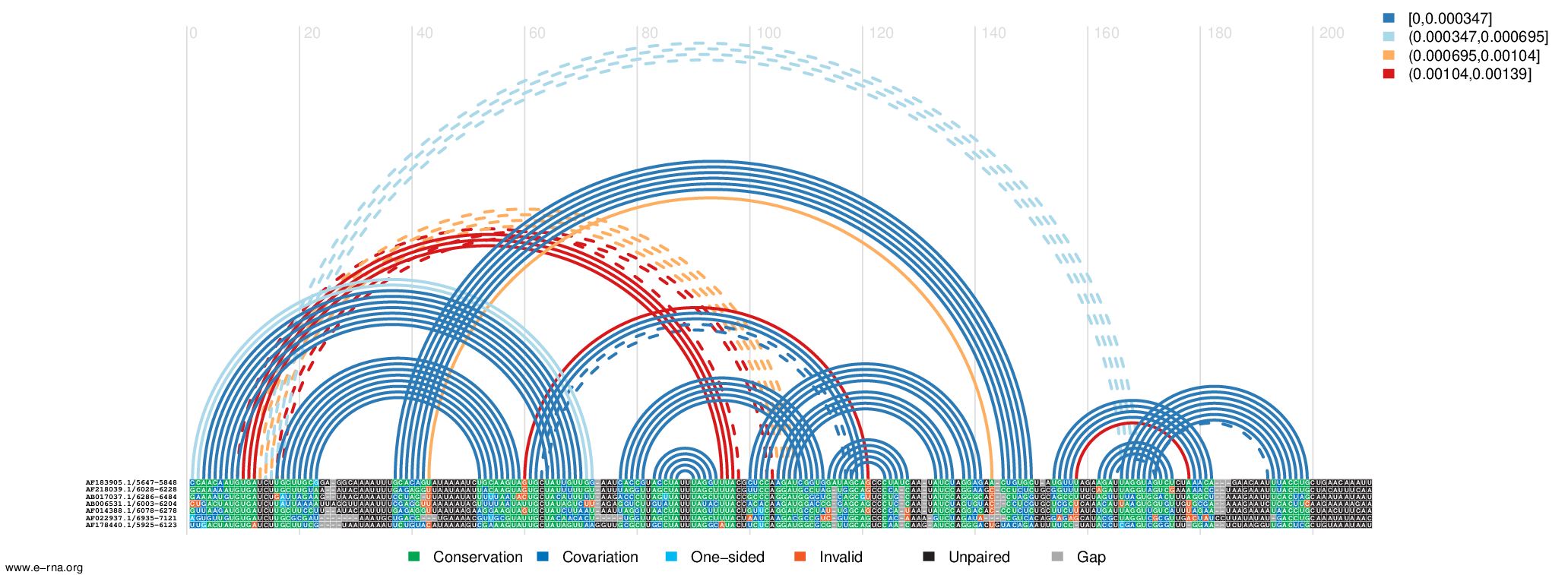
- Input: a multiple sequence alignment with a corresponding SHAPE reactivity profile and - optionally - a phylogenetic tree
- Output: predicted RNA secondary structure features
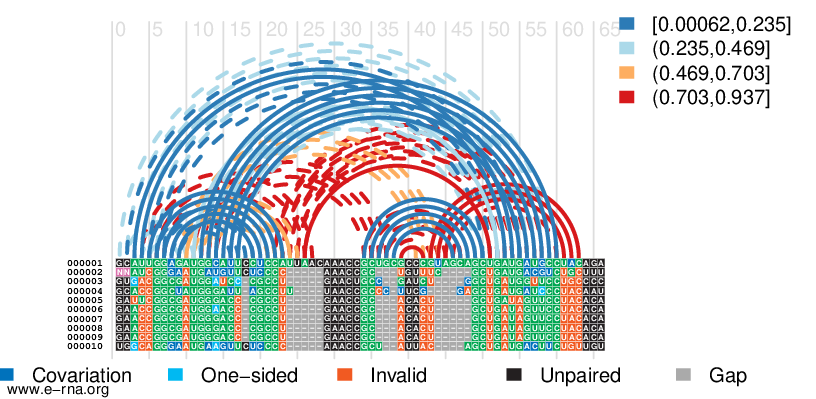
- Input: a set of un-aligned orthologous or homologous RNA sequences
- Output: the predicted consensus RNA secondary structure (potentially pseudo-knotted), a maximum a-posterior multiple sequence alignment, and the corresponding phylogenetic tree or network
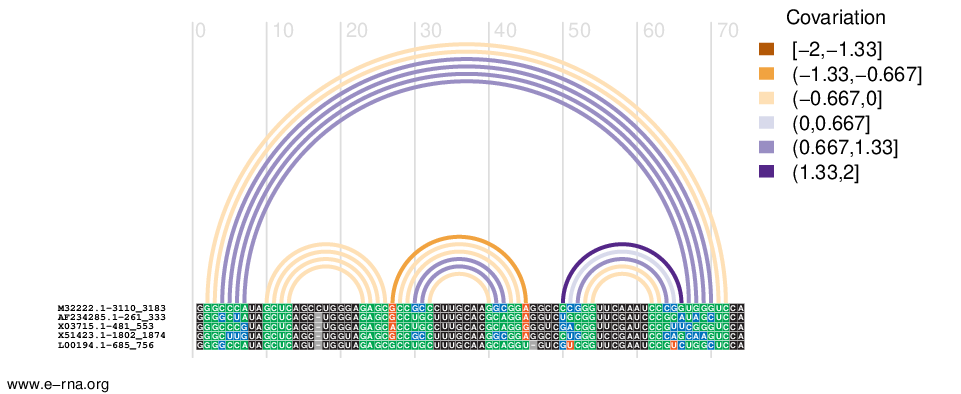
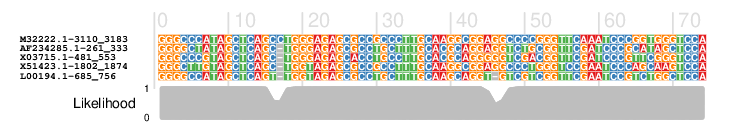
- Input: a multiple sequence alignment with annotated protein-coding regions and - optionally - a corresponding phylogenetic tree
- Output: predicted RNA secondary structures within input alignment
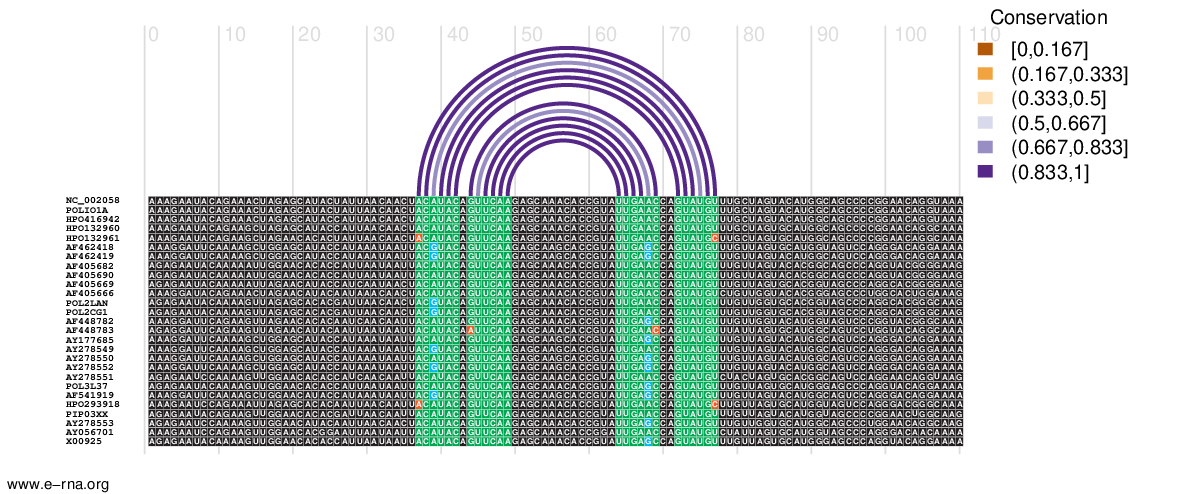
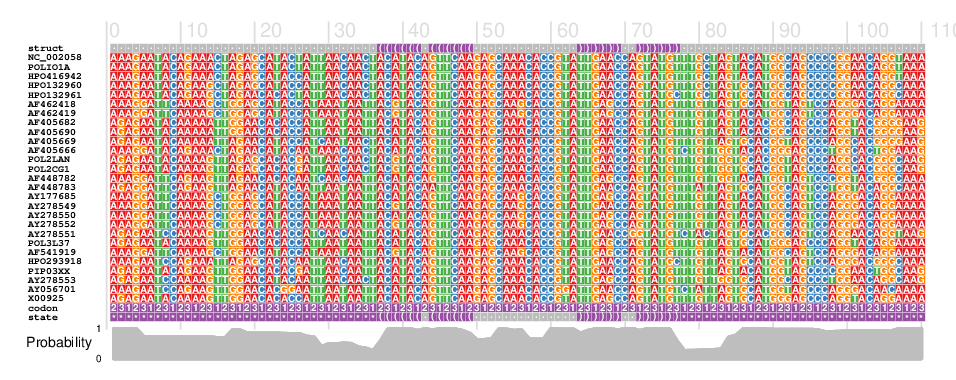
- Input: RNA sequence
- Output: predicted RNA secondary structure
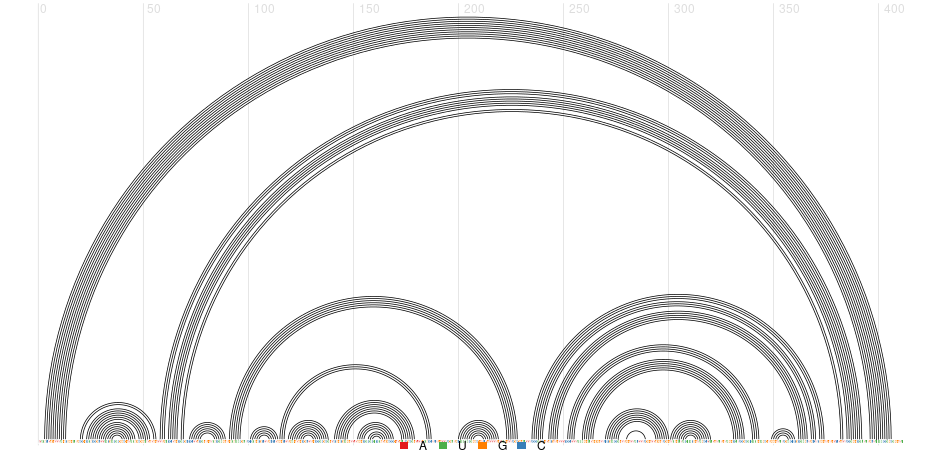
- Input: an RNA sequence or a corresponding multiple sequence alignment with a known reference RNA secondary structure
- Output: predicted RNA secondary structure features

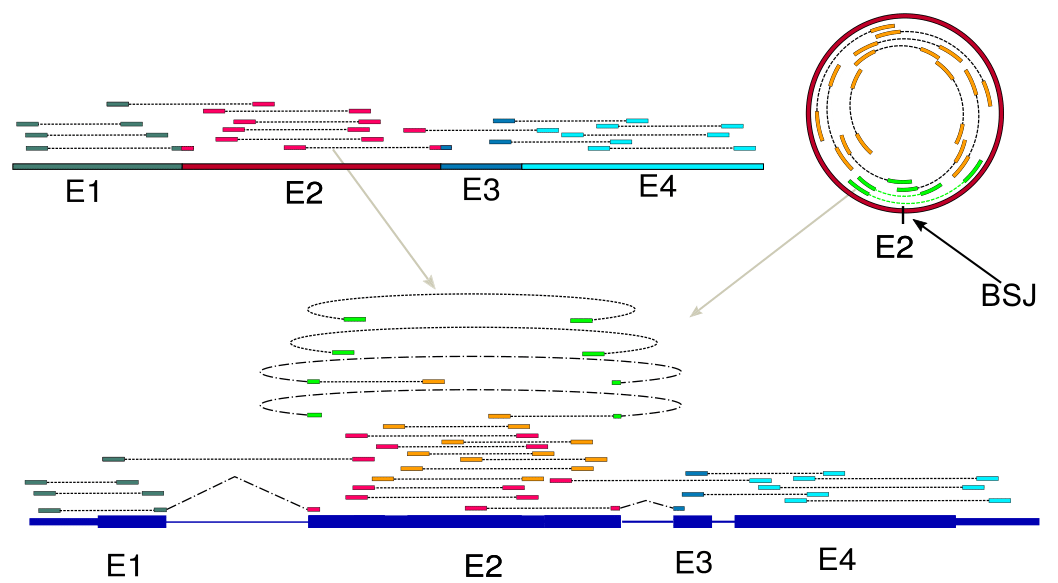
- Input: two RNA-seq libraries, one control and one circRNA-enriched RNA-seq library
- Output: predicted circRNAs, known linear isoforms and their respective expression levels
Licence
Web servers are available for everyone to use.
All tools are available to everyone under a CC BY-NC-ND licence. This means that users are only allowed to share (copy, distribute) for non-commercial purposes, but are not allowed to distribute adapted versions. Click to find out more about the licence.
Our supporters
We thank the following funding agencies for generously supporting our research.
Questions, comments, or problems? Contact us!
e-RNA.org | Meyer Lab | MDC Berlin
Last update on 19. December 2022.




